The ASReview team developed a procedure to overcome replication issues in creating a dataset for simulation studies to evaluate the performance of active learning models! In this blog post, we explain this procedure, called the “Noisy Label Filter (NLF) Procedure”.
Background: Labelled Data needed for a Simulation Study
In an ASReview simulation study, you can measure the performance of an active learning model using an already-labeled dataset containing the titles and abstracts of all the records reviewed during the screening phase, plus the labeling decisions. With this information, essentially, ASReview can simulate a scenario where your records haven’t been labeled yet, even though they have been. ASReview then mimics your labeling decisions (relevant/irrelevant), utilizing AI techniques to prioritize potentially relevant records. This simulation helps estimate the time an active learning model could save compared to manual screening or other models.
A fully labeled dataset is a prerequisite for a simulation study; ASReview cannot label your records without it, making a simulation impossible. Labelled dataset are available in the Synergy database, a free and open dataset on study selection in systematic reviews, comprising 169,288 academic works from 26 published systematic reviews. Alternatively, you can use your own dataset, for example, from an already published systematic review.
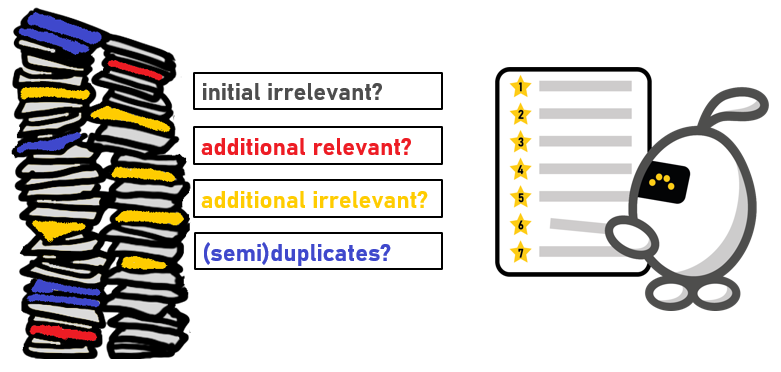
The problem: Noisy Labels
Unfortunately, obtaining a fully labeled dataset is often a challenge. Although sharing the search query has become the standard in the field, sharing the actual meta-data of all the records screened is not (i.e., titles and abstracts of the studies found in the search and all the labeling decisions of these records). To illustrate this point, we collected all systematic reviews published at Utrecht University and the University Medical Center in 2020. Out of 117 systematic reviews, 91 published the entire search query, but only 5 provided the actual underlying data. This is most likely caused because the PRISMA guidelines do not mandate researchers to archive their databases of included and excluded studies, but only the search query and the list of final inclusions. This raises a pressing question: How can we conduct a simulation study without a fully labeled dataset?
Well, one would think replicating the search query used to obtain the original dataset would result in exactly the same dataset, and one simply has to identify the relevant records in this set and label the remainder as irrelevant (see the ‘compose’ option in ASReview Datatools for an easy way to do this). Indeed, you can accurately label initially relevant records found in the reconstructed dataset, but it is unclear if the remainder of the records are actually irrelevant. This is because reproducing a search (many) years later might not result in exactly the same set of records: journals are added or omitted to databases, search functionalities change with new software releases, and the meta-data is continuously updated. Therefore, we do not know the exact record’s labels found by replicating the search because it is not guaranteed exactly the same records are part of the dataset
There are four possible types of records in the reconstructed dataset:
- Additional relevant records: These are records that were not part of the initial dataset, but should have been labeled as relevant when screened in the initial study. This is the most problematic type, as it leads to noisy labeled data for the simulation model. The presence of these records could confuse the model, causing incorrect categorization and unreliable results.
- Additional irrelevant records: These are new records found in the reconstructed search that should have been labeled as irrelevant in the initial study.
- Semi-duplicates of initial irrelevant records: These are records that were initially labeled irrelevant and deleted, but were left undeleted in the reconstructed database due to the de-duplication method.
- Initial irrelevant records: These are records that were initially labeled as irrelevant.
We call the labels of these records ‘noisy’. By labeling all noisy labels as irrelevant, we may falsely replicate the dataset! So, a second question arises: how can we replicate a systematic review without access to their complete list of labeling decisions?
The solution: NLF Procedure
So, recreating a fully labeled dataset that exactly matches the original might not feasible by ‘just’ replicating the search query for the reasons we discussed above. However, we can approximate the original dataset, including labeling decisions, as closely as possible, maximizing the utility of incomplete data in combination with screening a few records via ASReview.
Assuming we have replicated the search and obtained a list of records, the next step is to decide on the labels for those records that are not ‘proven’ to be relevant. Incorrect labeling of all these records as ‘irrelevant’ at this stage could result in a false replication of our dataset. To address this, we devised the Noisy Label Filter (NLF) Procedure.
The NLF procedure’s fundamental principle is to screen a small set of the most likely relevant records as predicted by an active learning model to determine if any noisy labels are relevant. Put simply, we ‘filter’ noisily labeled records into relevant and irrelevant. This way, with minimum screening effort, one can minimize the chances of mislabeling potentially relevant records as irrelevant.
How it works
- Reconstruct your dataset using initial search terms and check if the reconstructed dataset covers all initially relevant records.
- Then, find a way to label your initially relevant records as relevant in the reconstructed dataset. Use for example ASReview Datatools (script ‘compose’). If this step does not work, you could also manually select the relevant papers as prior knowledge in the project setup next step.
- Import the reconstructed dataset in ASReview Oracle and the relevant records will be recognized as prior knowledge. Also, select a set of random papers as irrelevant for completing the prior knowledge. If you have not labeled your relevant records yet, you can manually select all initially relevant papers as prior knowledge.
- Select your favorite model (or rely on the default settings).
- Using the initial inclusion and exclusion criteria screen a subset of the most likely relevant papers as predicted by the machine learning model. The screening can halt if 50 consecutive irrelevant papers are encountered, as finding any more relevant papers beyond this point is extremely unlikely.
- Upon completion, analyze any additional relevant records identified. Consider whether these records were omitted from the initial dataset or were mislabeled. Reflecting on these questions can provide insights into the discrepancies between the initial and reconstructed datasets.
- Assign the final labels to the dataset. The records you have not screened can be labeled as irrelevant since these are extremely unlikely to be relevant.
ASReview offers three different solutions to run simulations with the:
-
- Webapp (the frontend)
- Command line interface| (or use the Makita workflow generator)
- Python API
A Use-Case
To illustrate the NLF procedure, we used information from a systematic review of the psychological treatment of Borderline Personality Disorder (Oud et al., 2018).
Step 1: Prepare your Dataset
We replicated the search as precisely as possible using the following components:
- The search queries as reported by Oud et al., 2018.
- The number of papers identified for each search query, as reported in the search history file.
- Table 1 in Oud et al., covering initially included papers.
- The number of screened studies reported in the PRISMA flowchart in Oud et al. (i.e., n=1013).
In our paper, we explain the process in detail. A document including the initial search terms, the reconstructed search terms, and the number of results per query can be found on OSF.
The results of replicating the search are shown in Table 1. It is important to note that the information provided by the original researchers correctly followed the established PRISMA guidelines (even following the updated PRISMA guidelines, unpublished at the time of the paper). The reasons for the mismatch are mostly related to the black-box search engines, and we provide extensive explanations in the first section of our paper.
After exporting all the data from the search engines into the open-source reference manager Zotero, we collected a total of 1543 records.
Table 1: Overview of the number of queries whose results were exported to the reconstructed database
| Results in the initial query | Results in the reconstructed query | |
| PsycInfo | 533 | 566 |
| MEDLINE | 444 | 486 |
| CinaHL | 116 | 201 |
| Embase | 303 | 290 |
| Total | 1396 | 1543 |
We applied deduplication using ASReview Datatools followed by a manual deduplication of initially relevant records (see paper for more details).
Our version of the reconstructed database contained 1053 records. We verified that the initial inclusions were present in this database, which appeared to be the case. Next, we labeled 20 records as relevant based on the list of initial inclusions, with the exception of the initially included article by (Bateman & Fonagy, 1999), which was retracted after Oud et al.’s study was performed.
As said, our reconstructed dataset covered 1053 records whereas the original dataset covered 1013 records. The difference of 40 records between the initial study and our reconstructed database illustrates that these databases are not exactly the same. That is, we could find the initially included records in our reconstruction, but we don’t have the initially irrelevant records. That is why we are not sure about the other records in our reconstruction and we assign them noisy labels. By labeling all these records as irrelevant, we could miss additional relevant articles. These are records that were not part of the initial dataset but should have been labeled as relevant when screened in the initial study. This is problematic, as it leads to noisily labeled data for the simulation model. The presence of these records could confuse the model, causing incorrect categorization and unreliable results.
Step 2: Screen Noisy Labels in ASReview
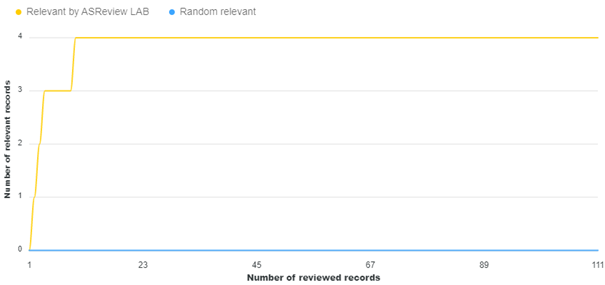
The NLF procedure was applied by the initial researcher, Matthijs Oud. As per the method section, the researcher imported the reconstructed dataset in v1.1 of ASReview, selected 20 papers as relevant (already labeled as relevant in the imported dataset), and pre-labeled five random records as irrelevant. During the screening phase, which lasted about an hour, the researcher labeled a total of 111 items, out of which four were additionally labeled as relevant. Figure 1 shows that these relevant items were found at the beginning of the first ten labels. The researcher later recalled that these additional relevant labels were initially not labeled as relevant because they seemed like relevant records to the researcher due to their proximity to initial relevant records or being follow-up studies.
Figure 1 Lapse of screening process in application NLF procedure
Step 3: Apply Final Labels
We made the decision to label all noisy labels as irrelevant, resulting in 1033 records being labeled as such, and the initially relevant studies were kept as relevant, except for the retracted paper. The irrelevant labels were added to the data via ASReview Datatools .
Step 4: Simulation Study
A simulation study was conducted using ASReview Makita. The ‘multiple models’ function was utilized to compare logistic regression, naïve Bayes, random forest, and support vector machine.
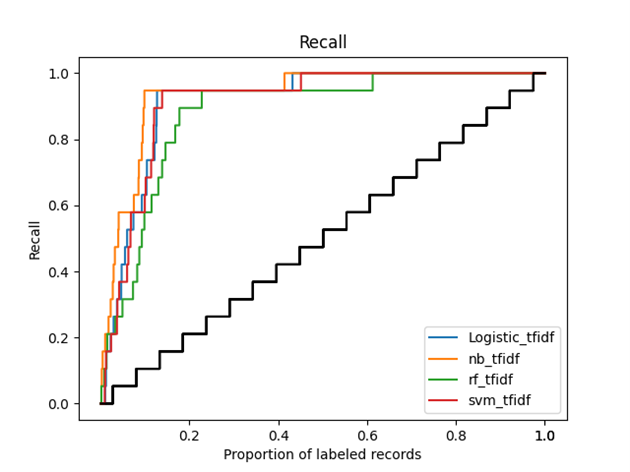
Figure 2 (recall curve) graphically shows the main outcomes of this simulation study. In the Figure, we can see that after screening around 10-20% of the total records, all active learning models found already about 95% of all relevant studies. The orange line (Naïve Bayes) is the first to reach 95% of included papers and the green line (Random Forest) is the last to reach that level.
Figure 2 Recall curve of the simulation study, with the x-axis representing the proportion of labeled records and the y-axis representing the proportion of already labeled relevant records. The black line represents the average process of manual screening, while the colored lines represent the performance of different active learning models in ASReview, with blue for Logistic Regression, orange for Naive Bayes, green for Random Forest, and red for Support Vector Machine.
To Cite
Neeleman, R. C., Leenaars, C., Oud, M., Weijdema, F., & van de Schoot, R. (2023, August 3). Addressing the Challenges of Reconstructing Systematic Reviews Datasets. DOI:10.31234/osf.io/jfcbq
If you have any questions, suggestions or additions to this procedure, we invite you to our GitHub-discussion board. You could contribute by further developing the NLF procedure!





